

Lukas Folkman
Lecturer in Information Technology
machine learning | data science | bioinformatics
Faculty of Science and Engineering
Southern Cross University, Australia

Lecturer in Information Technology
machine learning | data science | bioinformatics
Faculty of Science and Engineering
Southern Cross University, Australia
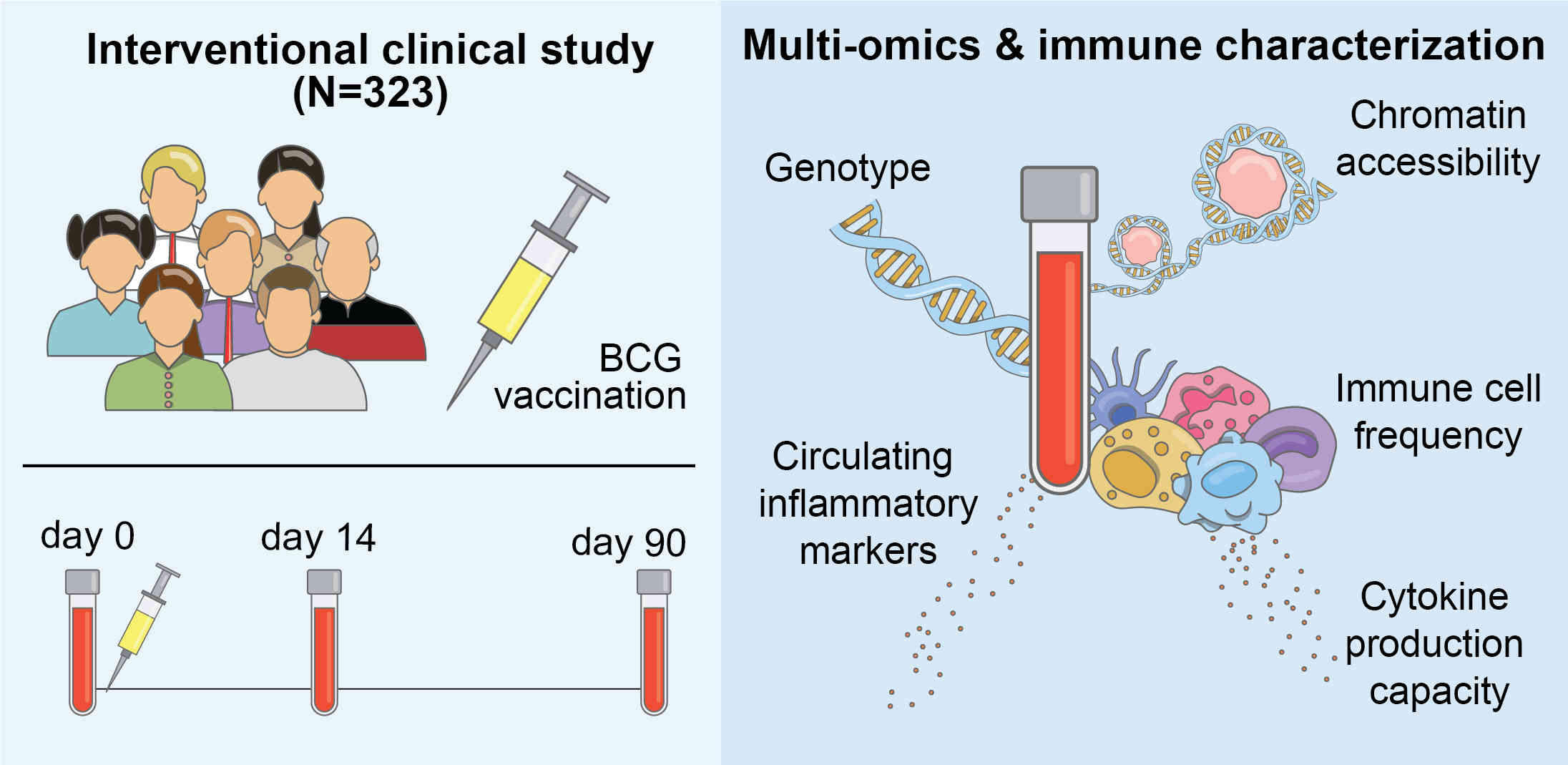
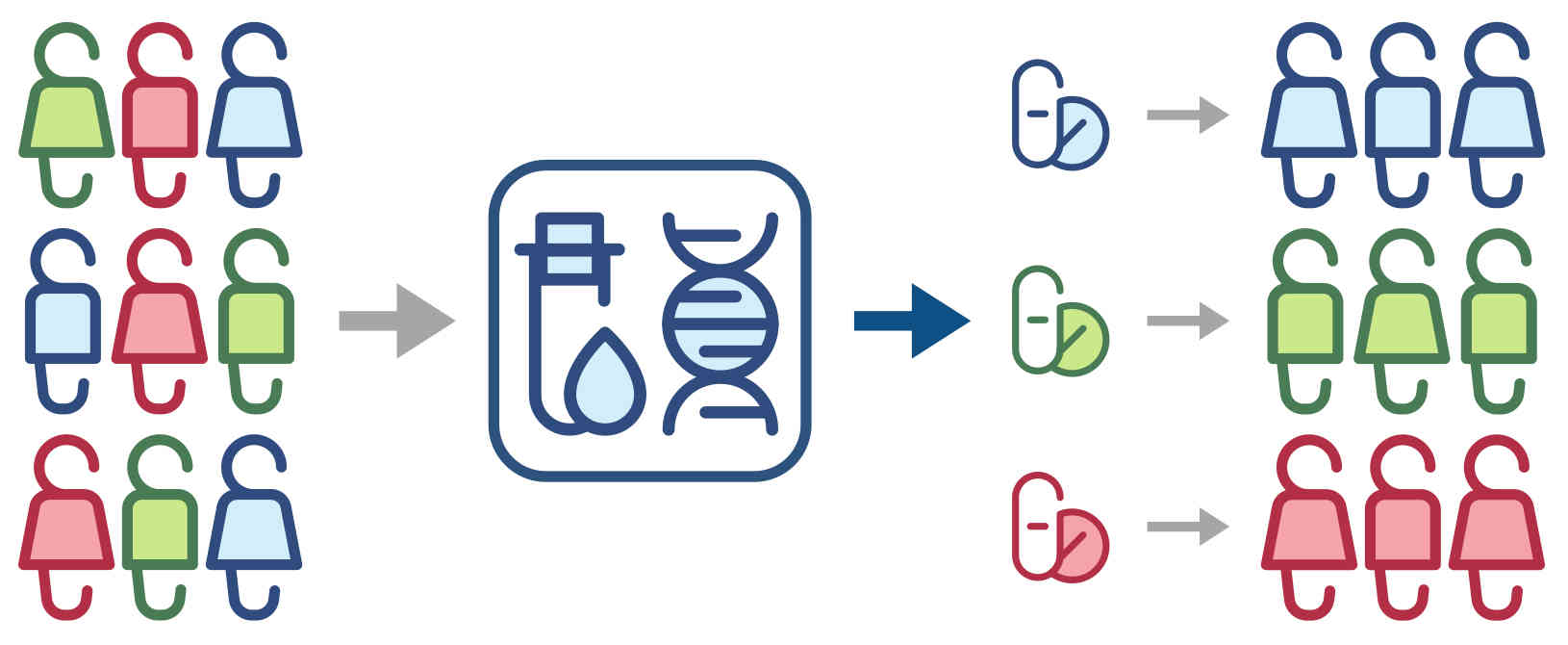
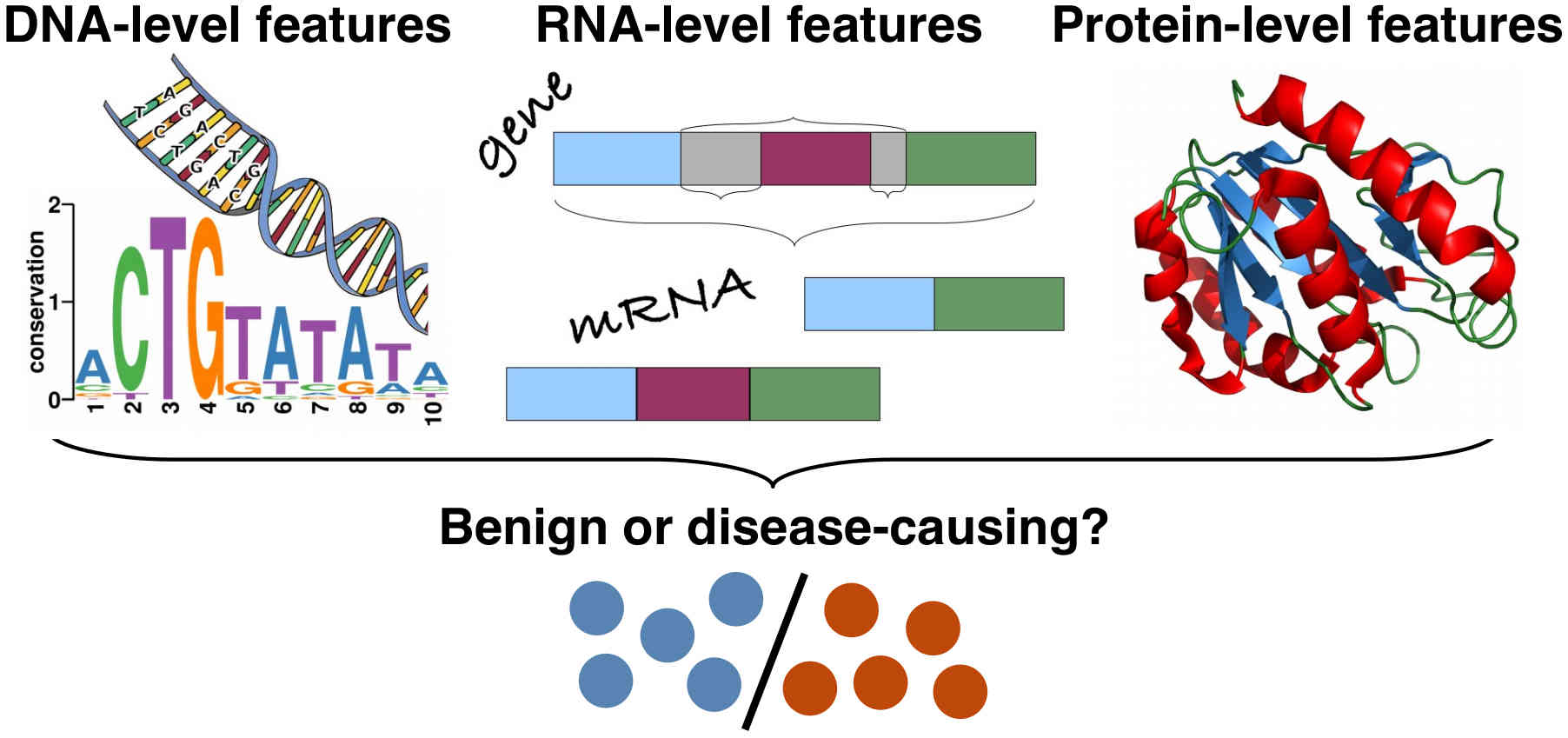
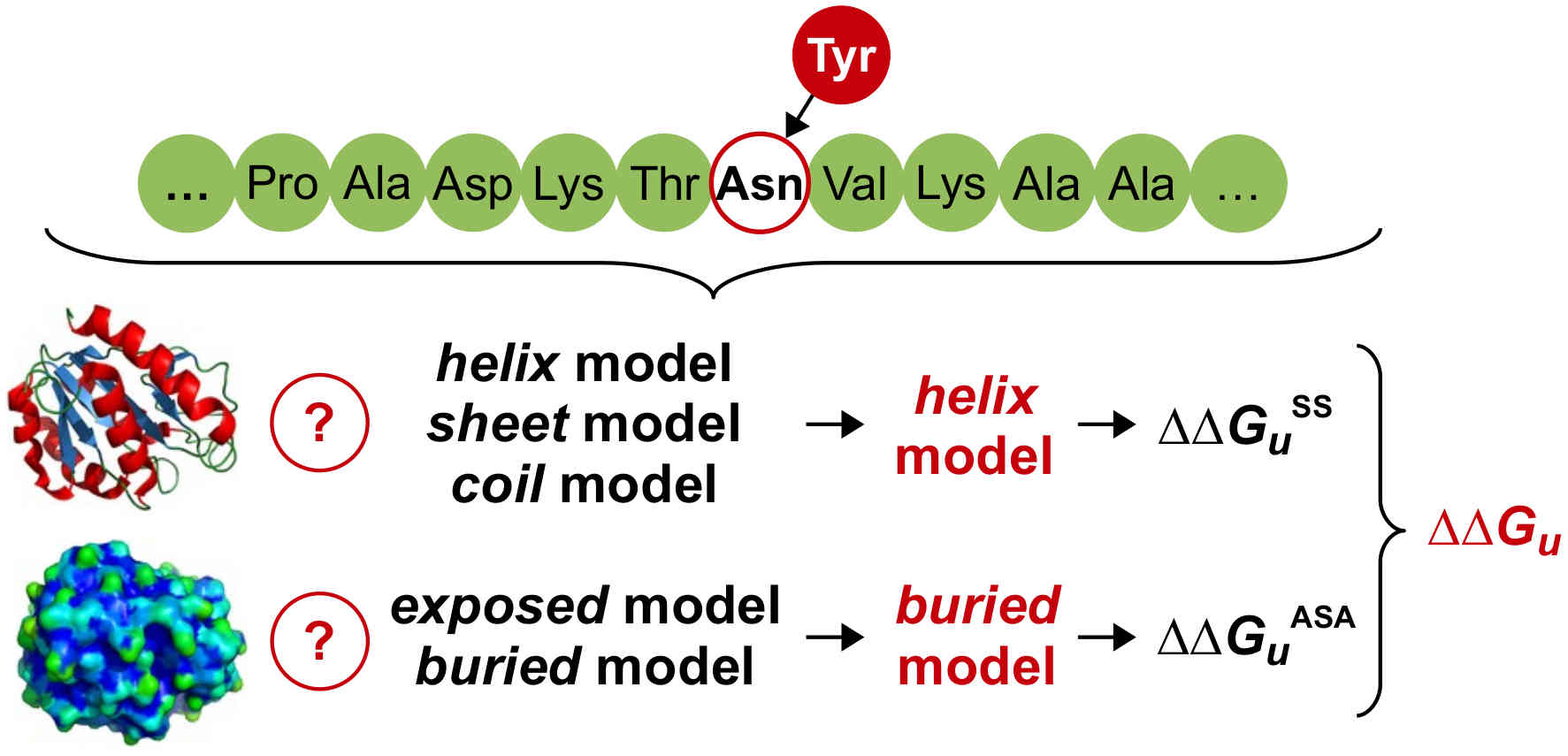
I am a computer scientist with research interests in developing machine learning methods for the life sciences. I earned my PhD from the Institute for Integrated and Intelligent Systems, Griffith University (Australia) in 2015, with a thesis on the prediction of stability and functional changes caused by genomic variants. Following this, I moved to Switzerland to continue my training as a postdoctoral fellow in the Machine Learning & Computational Biology Lab, ETH Zurich. There, my research focused on machine learning methods for precision medicine. In 2018, I moved to Austria to join CeMM Research Center for Molecular Medicine to develop machine learning methods for single-cell RNA-seq data analysis, for which I received European Commission's Marie Skłodowska-Curie Actions Postdoctoral Fellowship. While at CeMM, and later at the Institute of Artificial Intelligence, Medical University of Vienna, I was also the lead bioinformatician and data scientist for a project exploring epigenetic-immunological interplay and developed graph neural networks for virtual screening of small molecules. In 2022, I returned to Griffith University, where I worked as a Blue Economy Cooperative Research Centre research fellow at the Coastal and Marine Research Centre, developing computer vision methods for marine wildlife monitoring and precision aquaculture. Since 2025, I am a Lecturer (Assistant Professor) in Information Technology at Faculty of Science and Engineering, Southern Cross University, Australia.


# equal contributions * senior author(s)