|
Publications
# denotes equal contributions
* denotes senior author(s)
-
Folkman L, Vo QLK, Johnston C, Stantic B, Pitt KA (2025)
A computer vision method to estimate ventilation rate of Atlantic salmon in sea fish farms
Aquacultural Engineering, 112, 102645
📄 [Open access]
-
Vo QLK, Pitt KA, Johnston C, Kennedy B, Folkman* L (2025)
Computer vision detects an association between gross gill score and ventilation rates in farmed Atlantic salmon (Salmo salar)
Journal of Fish Diseases, e70055
📄 [Open access]
-
Folkman L, Pitt KA, Stantic B (2025)
A data-centric framework for combating domain shift in underwater object detection with image enhancement
Applied Intelligence 55, 272
📄 [Open access] [GitHub]
-
Traxler# P, Reichl# S, Folkman L, …, Farlik* M, Bock* C (2025)
Integrated time-series analysis and high-content CRISPR screening delineate the dynamics of macrophage immune regulation
Cell Systems 16, 101346
📄 [Open access] [Press release] [Website] [GitHub]
-
Teufel LU, Matzaraki V, Folkman L, ..., Arts RJW (2025)
Interleukin-38 is a negative regulator of trained im- munity: A retrospective multi-omics study
iScience, 28(11), 113758
📄 [Open access]
-
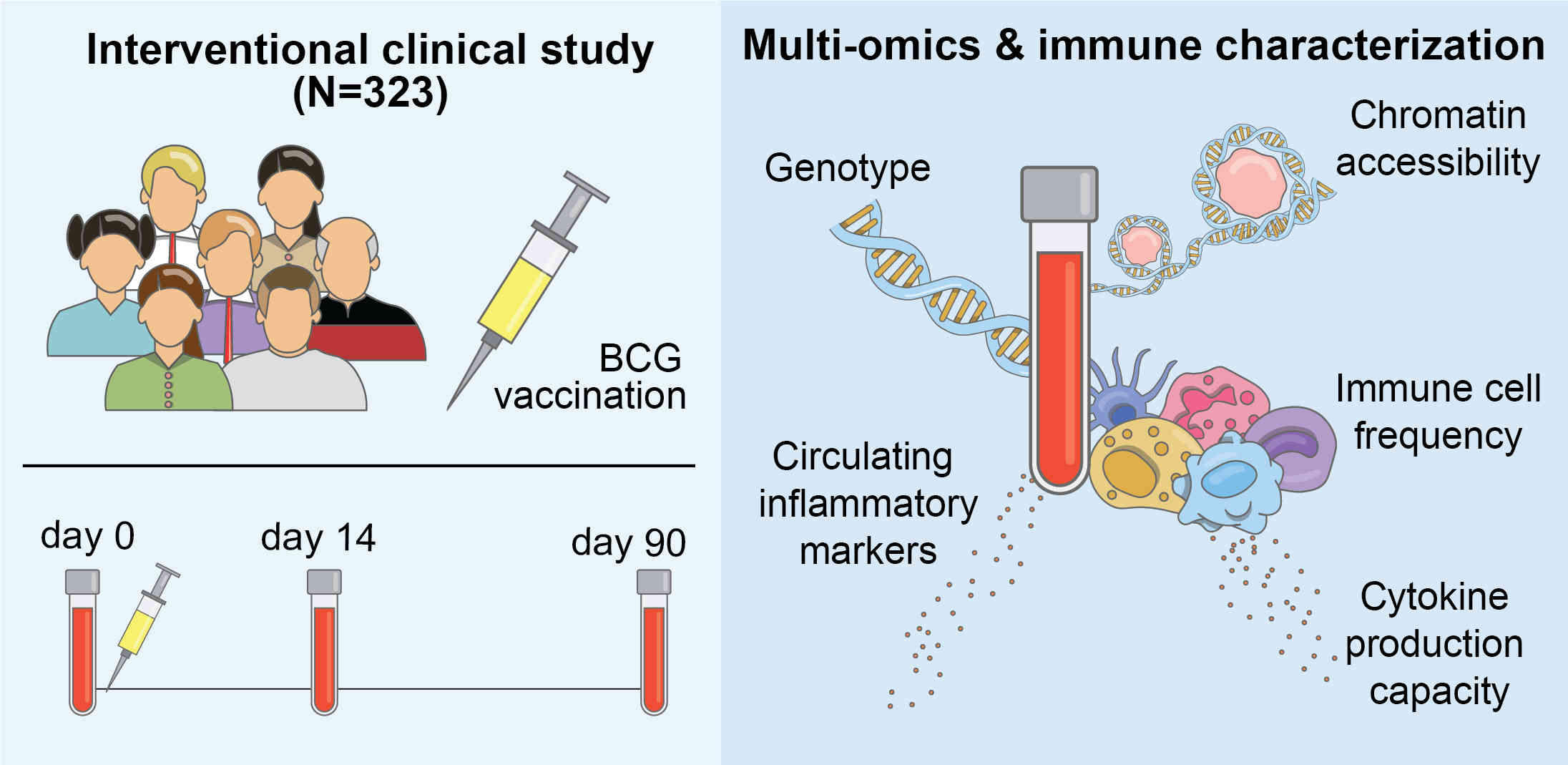
Moorlag# SJCFM, Folkman# L, ter Horst# R, Krausgruber# T, Barreca B, Schuster LC, Fife V, Matzaraki V, Li W, Reichl S, Mourits VP, Koeken VACM, de Bree LCJ, Dijkstra H, Lemmers H, van Cranenbroek B, van Rijssen E, Koenen HJPM, Joosten I, Xu C,Li Y, Joosten LAB, van Crevel R, Netea* MG & Bock* C (2024)
Multi-omics analysis of innate and adaptive responses to BCG vaccination reveals epigenetic cell states that predict trained immunity
Immunity 57(1), 171–187
📄 [Open access] [PubMed] [Press release] [Website] [GitHub]
★ Selected for Nature Research Highlights
-
Teufel LU, Matzaraki V, Folkman L, ter Horst R, Moorlag SJCFM, Mulders-Manders CM, Netea MG, Krausgruber T, Joosten LAB, Arts RJW (2024)
Insights into the multifaceted role of interleukin-37 on human immune cell regulation
Clinical Immunology 268(1), 110368
📄 [Open access] [PubMed]
-
Turina# P, Cortivo# GD, …, Folkman L, …, Dell’Orco* D & Capriotti* E (2025)
Assessing the predicted impact of single amino acid substitutions in calmodulin for CAGI6 challenges
Human Genetics 144, 113–125
📄 [Article] [PubMed]
-
Turina# P, Petrosino# M, …, Folkman L, …, Chiaraluce* R, Consalvi* V & Capriotti* E (2025)
Assessing the predicted impact of single amino acid substitution in MAPK proteins for CAGI6 challenges
Human Genetics 144, 265–280
📄 [Article] [PubMed]
-
The Critical Assessment of Genome Interpretation Consortium (2024)
CAGI, the Critical Assessment of Genome Interpretation, establishes progress and prospects for computational genetic variant interpretation methods
Genome Biology 25, 53
📄 [Open access] [PubMed]
-
de Bree LCJ, Mourits VP, Koeken VACM, Moorlag SJCFM, Janssen R, Folkman L, Barreca D, Krausgruber T, Fife-Gernedl V, Novakovic B, Arts RJW, Dijkstra H, Lemmers H, Bock C, Joosten LAB, van Crevel R, Benn CS & Netea MG (2020)
Circadian rhythm influences induction of trained immunity by BCG vaccination
The Journal of Clinical Investigation 130(10), 5603–5617
📄 [Open access] [PubMed]
-
Pejaver V, Babbi G, Casadio R, Folkman L, Katsonis P, Kundu K, Lichtarge O, Martelli PL, Miller M, Moult J, Pal LR, Savojardo C, Yin Y, Zhou Y, Radivojac P & Bromberg Y (2019)
Assessment of methods for predicting the effects of PTEN and TPMT protein variants
Human Mutation 40(9), 1495–1506
📄 [Article] [Open access via PMC] [PubMed] [CAGI5]
-
Savojardo C, Petrosino M, Babbi G, Bovo S, Corbi‐Verge C, Casadio R, Fariselli P, Folkman L, Garg A, Karimi M, Katsonis P, Kim PM, Lichtarge O, Martelli PL, Pasquo A, Pal D, Shen Y, Strokach AV, Turina P, Zhou Y, Andreoletti G, Brenner S, Chiaraluce R, Consalvi V & Capriotti E (2019)
Evaluating the predictions of the protein stability change upon single amino acid substitutions for the FXN CAGI5 challenge
Human Mutation 40(9), 1392–1399
📄 [Article] [Open access via PMC] [PubMed] [CAGI5]
-
Clark WT, Kasak L, Bakolitsa C, Hu Z, Andreoletti G, Babbi G, Bromberg Y, Casadio R, Dunbrack R, Folkman L, Ford CT, Jones D, Katsonis P, Kundu K, Lichtarge O, Martelli PL, Mooney SD, Nodzak C, Pal LR, Radivojac P, Savojardo C, Shi X, Zhou Y, Uppal A, Xu Q, Yin Y, Pejaver V, Wang M, Wei L, Moult J, Yu GK, Brenner SE & LeBowitz JH (2019)
Assessment of predicted enzymatic activity of alpha‐N‐acetylglucosaminidase (NAGLU) variants of unknown significance for CAGI 2016
Human Mutation 40(9), 1519–1529
📄 [Article] [Open access via PMC] [PubMed] [CAGI4]
-
He# X, Folkman# L & Borgwardt K (2018)
Kernelized rank learning for personalized drug recommendation
Bioinformatics 34(16), 2808–2816
📄 [Open access] [Poster] [PubMed] [GitHub] [Datasets]
★ ISMB/ECCB 2017 and [BC]2 2017 best poster awards
-
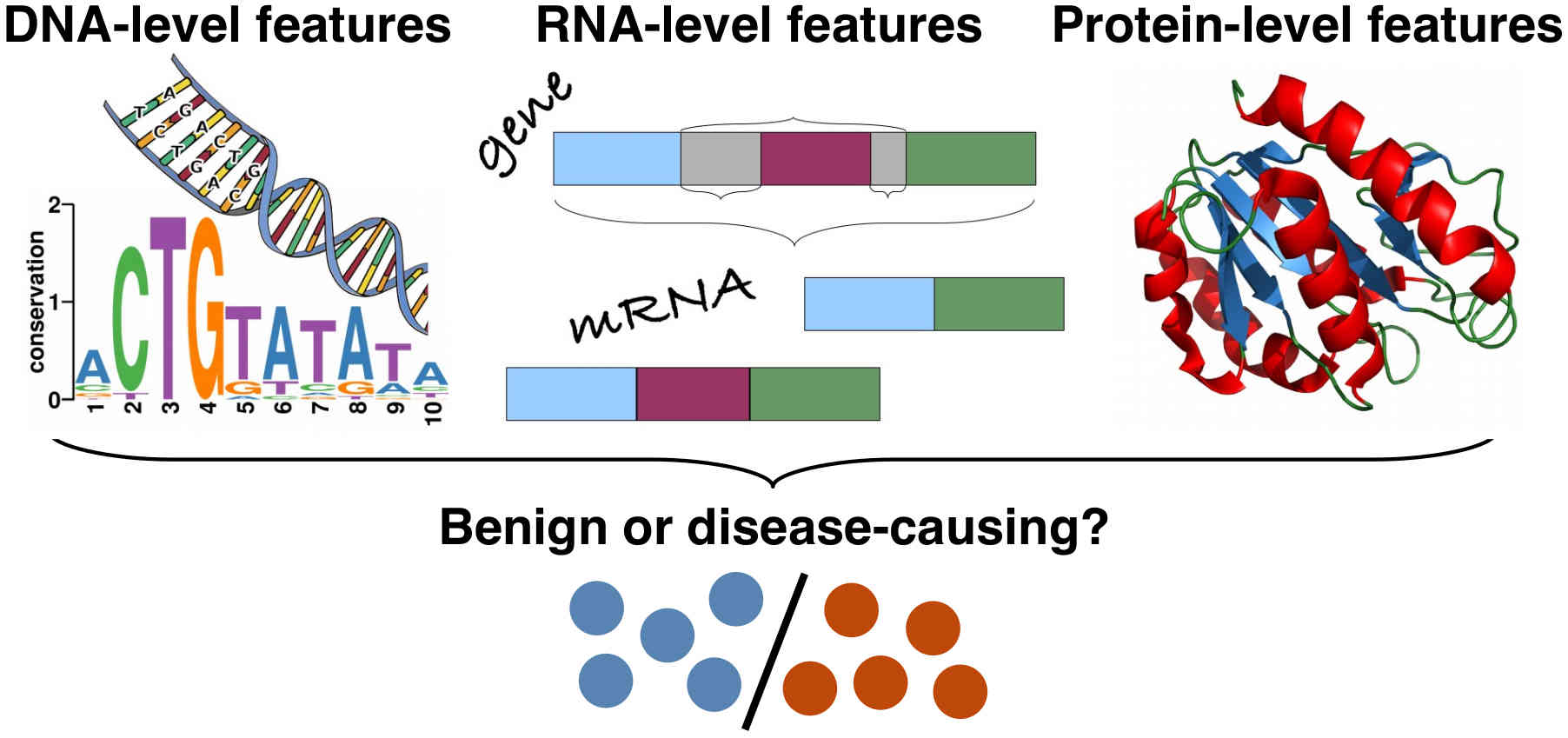
Livingstone# M, Folkman# L, Yang Y, Zhang P, Mort M, Cooper DN, Liu Y, Stantic B & Zhou Y (2017)
Investigating DNA-, RNA-, and protein-based features as a means to discriminate pathogenic synonymous variants
Human Mutation 38(10), 1336–1347
📄 [Article] [Preprint] [PubMed] [Web server and datasets] [Singularity container]
-
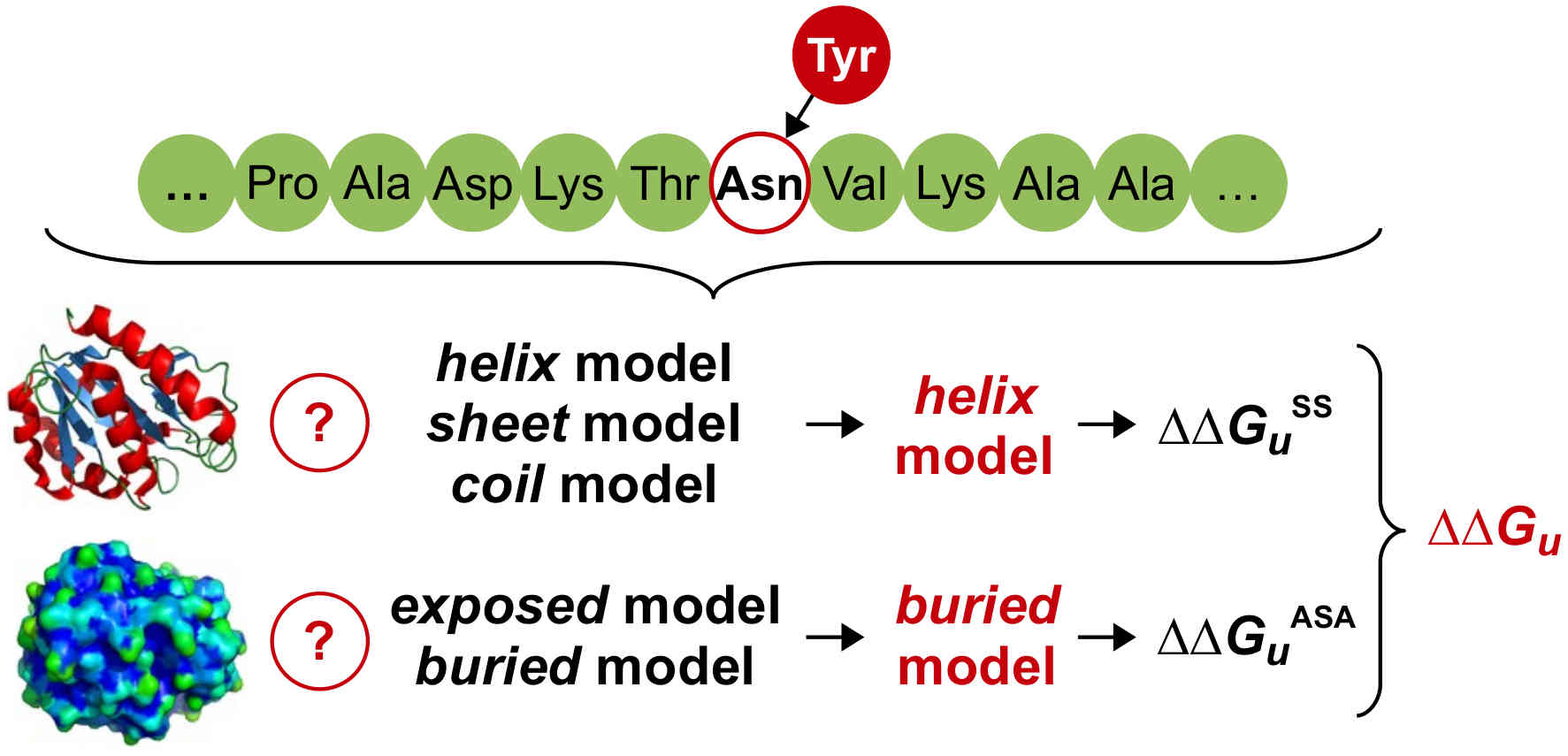
Folkman L, Stantic B, Sattar A & Zhou Y (2016)
EASE-MM: sequence-based prediction of mutation-induced stability changes with feature-based multiple models
Journal of Molecular Biology, 428(6), 1394–1405
📄 [Article] [Preprint] [PubMed] [Web server and datasets] [Singularity container]
★ Ranked 3rd in the frataxin stability prediction during the Critical Assessment of Genome Interpretation 2018 competition
-
Folkman L, Yang Y, Li Z, Stantic B, Sattar B, Mort M, Cooper DN, Liu Y & Zhou Y (2015)
DDIG-in: detecting disease-causing genetic variations due to frameshifting indels and nonsense mutations by sequence and structural properties at nucleotide and protein levels
Bioinformatics 31(10), 1599–1606
📄 [Open access] [PubMed] [Web server and datasets] [Singularity container]
★ Winner of the Three Minutes Thesis Competition 2014 at the School of Information and Communication Technology, Griffith University
-
Folkman L, Stantic B & Sattar A (2014)
Feature-based multiple models improve classification of mutation-induced stability changes
BMC Genomics 15(Suppl 4), S6
📄 [Open access] [PubMed]
-
Folkman L, Stantic B & Sattar A (2014)
Towards sequence-based prediction of mutation-induced stability changes in unseen non-homologous proteins
BMC Genomics 15(Suppl 1), S4
📄 [Open access] [PubMed]
-
Folkman L, Stantic B & Sattar A (2013)
Sequence-only evolutionary and predicted structural features for the prediction of stability changes in protein mutants
BMC Bioinformatics 14(Suppl 2), S6
📄 [Open access] [PubMed]
-
Higgs T, Folkman L & Stantic B (2013)
Combining protein fragment feature-based resampling and local optimisation
in ‘IAPR International Conference on Pattern Recognition in Bioinformatics (PRIB)’, Vol. 7986 of LNCS, pp. 114–125, Springer
📄 [Article]
-
Folkman L, Pullan W & Stantic B (2011)
Generic parallel genetic algorithm framework for protein optimisation
in ‘Algorithms and architectures for parallel processing (ICA3PP)’, Vol. 7017 of LNCS, pp. 64–73, Springer
📄 [Article]
-
Stetsko A, Folkman L, & Matyas V (2010)
Neighbor-based intrusion detection in wireless sensor networks
in ‘Wireless and Mobile Communications (ICWMC)’, pp. 420–42, IEEE
📄 [Article]
|